Profile Likelihood Plots for 'rma' Objects
profile.rma.RdFunctions to profile the (restricted) log-likelihood for objects of class "rma.uni", "rma.mv", "rma.uni.selmodel", and "rma.ls".
# S3 method for class 'rma.uni'
profile(fitted, xlim, ylim, steps=20, lltol=1e-03,
progbar=TRUE, parallel="no", ncpus=1, cl, plot=TRUE, ...)
# S3 method for class 'rma.mv'
profile(fitted, sigma2, tau2, rho, gamma2, phi, xlim, ylim, steps=20, lltol=1e-03,
progbar=TRUE, parallel="no", ncpus=1, cl, plot=TRUE, ...)
# S3 method for class 'rma.uni.selmodel'
profile(fitted, tau2, delta, xlim, ylim, steps=20, lltol=1e-03,
progbar=TRUE, parallel="no", ncpus=1, cl, plot=TRUE, ...)
# S3 method for class 'rma.ls'
profile(fitted, alpha, xlim, ylim, steps=20, lltol=1e-03,
progbar=TRUE, parallel="no", ncpus=1, cl, plot=TRUE, ...)
# S3 method for class 'profile.rma'
print(x, ...)
# S3 method for class 'profile.rma'
plot(x, xlim, ylim, pch=19, xlab, ylab, main, refline=TRUE, cline=FALSE, ...)Arguments
- fitted
an object of class
"rma.uni","rma.mv","rma.uni.selmodel", or"rma.ls".- x
an object of class
"profile.rma"(forplotandprint).- sigma2
optional integer to specify for which \(\sigma^2\) parameter the likelihood should be profiled.
- tau2
optional integer to specify for which \(\tau^2\) parameter the likelihood should be profiled.
- rho
optional integer to specify for which \(\rho\) parameter the likelihood should be profiled.
- gamma2
optional integer to specify for which \(\gamma^2\) parameter the likelihood should be profiled.
- phi
optional integer to specify for which \(\phi\) parameter the likelihood should be profiled.
- delta
optional integer to specify for which \(\delta\) parameter the likelihood should be profiled.
- alpha
optional integer to specify for which \(\alpha\) parameter the likelihood should be profiled.
- xlim
optional vector to specify the lower and upper limit of the parameter over which the profiling should be done. If unspecified, the function sets these limits automatically.
- ylim
optional vector to specify the y-axis limits when plotting the profiled likelihood. If unspecified, the function sets these limits automatically.
- steps
number of points between
xlim[1]andxlim[2](inclusive) for which the likelihood should be evaluated (the default is 20). Can also be a numeric vector of length 2 or longer to specify for which parameter values the likelihood should be evaluated (in this case,xlimis automatically set torange(steps)if unspecified).- lltol
numerical tolerance used when comparing values of the profiled log-likelihood with the log-likelihood of the fitted model (the default is 1e-03).
- progbar
logical to specify whether a progress bar should be shown (the default is
TRUE).- parallel
character string to specify whether parallel processing should be used (the default is
"no"). For parallel processing, set to either"snow"or"multicore". See ‘Details’.- ncpus
integer to specify the number of processes to use in the parallel processing.
- cl
optional cluster to use if
parallel="snow". If unspecified, a cluster on the local machine is created for the duration of the call.- plot
logical to specify whether the profile plot should be drawn after profiling is finished (the default is
TRUE).- pch
plotting symbol to use. By default, a filled circle is used. See
pointsfor other options.- refline
logical to specify whether the value of the parameter estimate should be indicated by a dotted vertical line and its log-likelihood value by a dotted horizontal line (the default is
TRUE).- cline
logical to specify whether a horizontal reference line should be added to the plot that indicates the log-likelihood value corresponding to the 95% profile confidence interval (the default is
FALSE). Can also be a numeric value between 0 and 100 to specify the confidence interval level.- xlab
title for the x-axis. If unspecified, the function sets an appropriate axis title.
- ylab
title for the y-axis. If unspecified, the function sets an appropriate axis title.
- main
title for the plot. If unspecified, the function sets an appropriate title.
- ...
other arguments.
Details
The function fixes a particular parameter of the model and then computes the maximized (restricted) log-likelihood over the remaining parameters of the model. By fixing the parameter of interest to a range of values, a profile of the (restricted) log-likelihood is constructed.
Selecting the Parameter(s) to Profile
The parameters that can be profiled depend on the model object:
For objects of class
"rma.uni"obtained with therma.unifunction, the function profiles over \(\tau^2\) (not for equal-effects models).For objects of class
"rma.mv"obtained with therma.mvfunction, profiling is done by default over all variance and correlation components of the model. Alternatively, one can use thesigma2,tau2,rho,gamma2, orphiarguments to specify over which parameter the profiling should be done. Only one of these arguments can be used at a time. A single integer is used to specify the number of the parameter.For selection model objects of class
"rma.uni.selmodel"obtained with theselmodelfunction, profiling is done by default over \(\tau^2\) (for models where this is an estimated parameter) and all selection model parameters. Alternatively, one can choose to profile only \(\tau^2\) by settingtau2=TRUEor one can select one of the selection model parameters to profile by specifying its number via thedeltaargument.For location-scale model objects of class
"rma.ls"obtained with therma.unifunction, profiling is done by default over all \(\alpha\) parameters that are part of the scale model. Alternatively, one can select one of the parameters to profile by specifying its number via thealphaargument.
Interpreting Profile Likelihood Plots
A profile likelihood plot should show a single peak at the corresponding ML/REML estimate. If refline=TRUE (the default), the value of the parameter estimate is indicated by a dotted vertical line and its log-likelihood value by a dotted horizontal line. Hence, the intersection of these two lines should correspond to the peak (assuming that the model was fitted with ML/REML estimation).
When profiling a variance component (or some other parameter that cannot be negative), the peak may be at zero (if this corresponds to the ML/REML estimate of the parameter). In this case, the profiled log-likelihood should be a monotonically decreasing function of the parameter. Similarly, when profiling a correlation component, the peak may be at -1 or +1.
If the profiled log-likelihood has multiple peaks, this indicates that the likelihood surface is not unimodal. In such cases, the ML/REML estimate may correspond to a local optimum (when the intersection of the two dotted lines is not at the highest peak).
If the profile is flat (over the entire parameter space or large portions of it), then this suggests that at least some of the parameters of the model are not identifiable (and the parameter estimates obtained are to some extent arbitrary). See Raue et al. (2009) for some further discussion of parameter identifiability and the use of profile likelihoods to check for this.
The function checks whether any profiled log-likelihood value is actually larger than the log-likelihood of the fitted model (using a numerical tolerance of lltol). If so, a warning is issued as this might indicate that the optimizer did not identify the actual ML/REML estimate of the parameter profiled.
Parallel Processing
Profiling requires repeatedly refitting the model, which can be slow when \(k\) is large and/or the model is complex (the latter especially applies to "rma.mv" objects and also to certain "rma.uni.selmodel" or "rma.ls" objects). On machines with multiple cores, one can try to speed things up by delegating the model fitting to separate worker processes, that is, by setting parallel="snow" or parallel="multicore" and ncpus to some value larger than 1. Parallel processing makes use of the parallel package, using the makePSOCKcluster and parLapply functions when parallel="snow" or using mclapply when parallel="multicore" (the latter only works on Unix/Linux-alikes).
Value
An object of class "profile.rma". The object is a list (or list of such lists) containing the following components:
One of the following (depending on the parameter that was actually profiled):
- sigma2
values of \(\sigma^2\) over which the likelihood was profiled.
- tau2
values of \(\tau^2\) over which the likelihood was profiled.
- rho
values of \(\rho\) over which the likelihood was profiled.
- gamma2
values of \(\gamma^2\) over which the likelihood was profiled.
- phi
values of \(\phi\) over which the likelihood was profiled.
- delta
values of \(\delta\) over which the likelihood was profiled.
- alpha
values of \(\alpha\) over which the likelihood was profiled.
In addition, the following components are included:
- ll
(restricted) log-likelihood values at the corresponding parameter values.
- beta
a matrix with the estimated model coefficients at the corresponding parameter values.
- ci.lb
a matrix with the lower confidence interval bounds of the model coefficients at the corresponding parameter values.
- ci.ub
a matrix with the upper confidence interval bounds of the model coefficients at the corresponding parameter values.
- ...
some additional elements/values.
Note that the list is returned invisibly.
References
Raue, A., Kreutz, C., Maiwald, T., Bachmann, J., Schilling, M., Klingmuller, U., & Timmer, J. (2009). Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics, 25(15), 1923–1929. https://doi.org/10.1093/bioinformatics/btp358
Viechtbauer, W. (2010). Conducting meta-analyses in R with the metafor package. Journal of Statistical Software, 36(3), 1–48. https://doi.org/10.18637/jss.v036.i03
Viechtbauer, W., & López-López, J. A. (2022). Location-scale models for meta-analysis. Research Synthesis Methods. 13(6), 697–715. https://doi.org/10.1002/jrsm.1562
See also
Examples
### calculate log odds ratios and corresponding sampling variances
dat <- escalc(measure="OR", ai=tpos, bi=tneg, ci=cpos, di=cneg, data=dat.bcg)
### fit random-effects model using rma.uni()
res <- rma(yi, vi, data=dat)
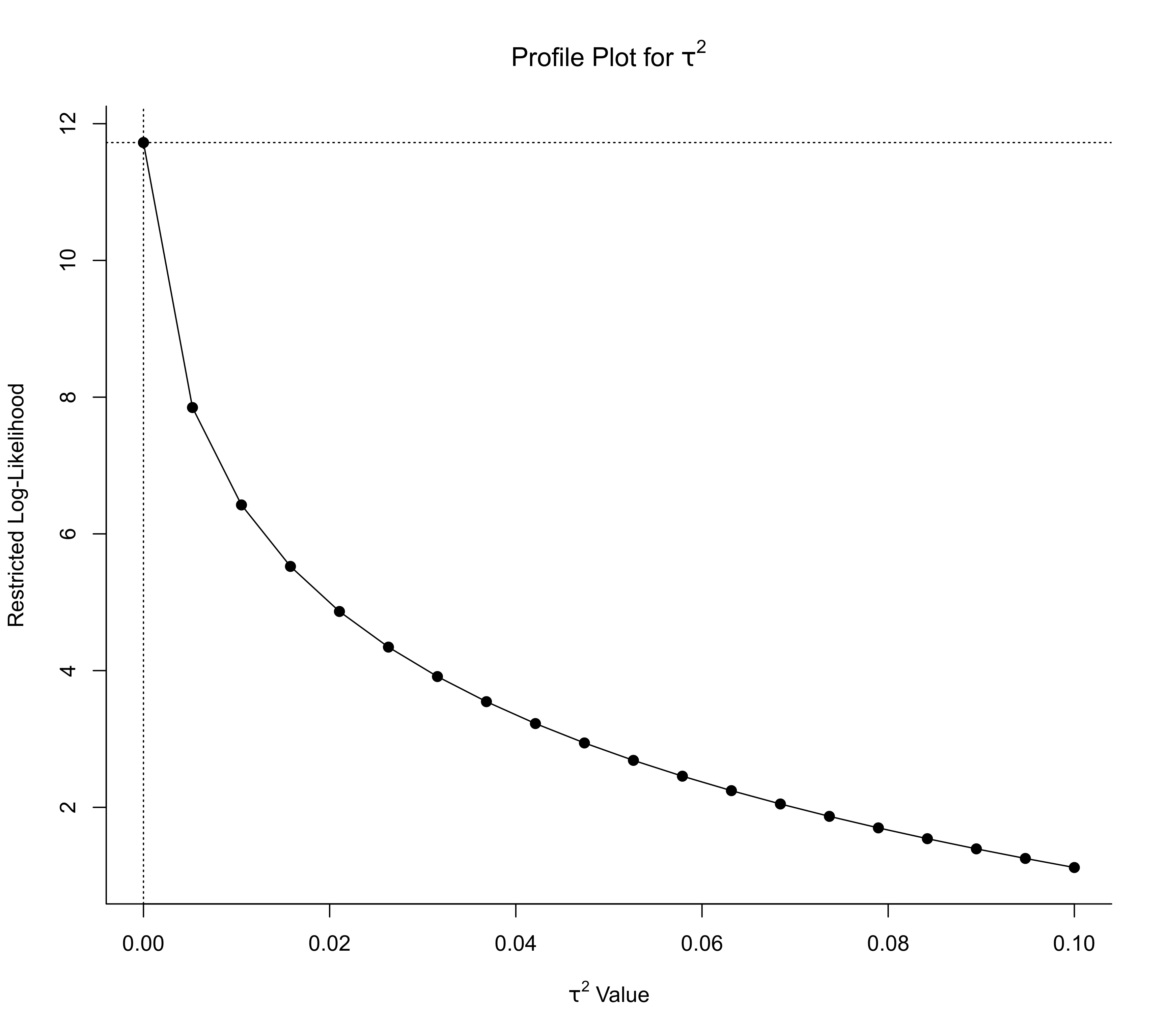
### profile over tau^2
profile(res, progbar=FALSE)
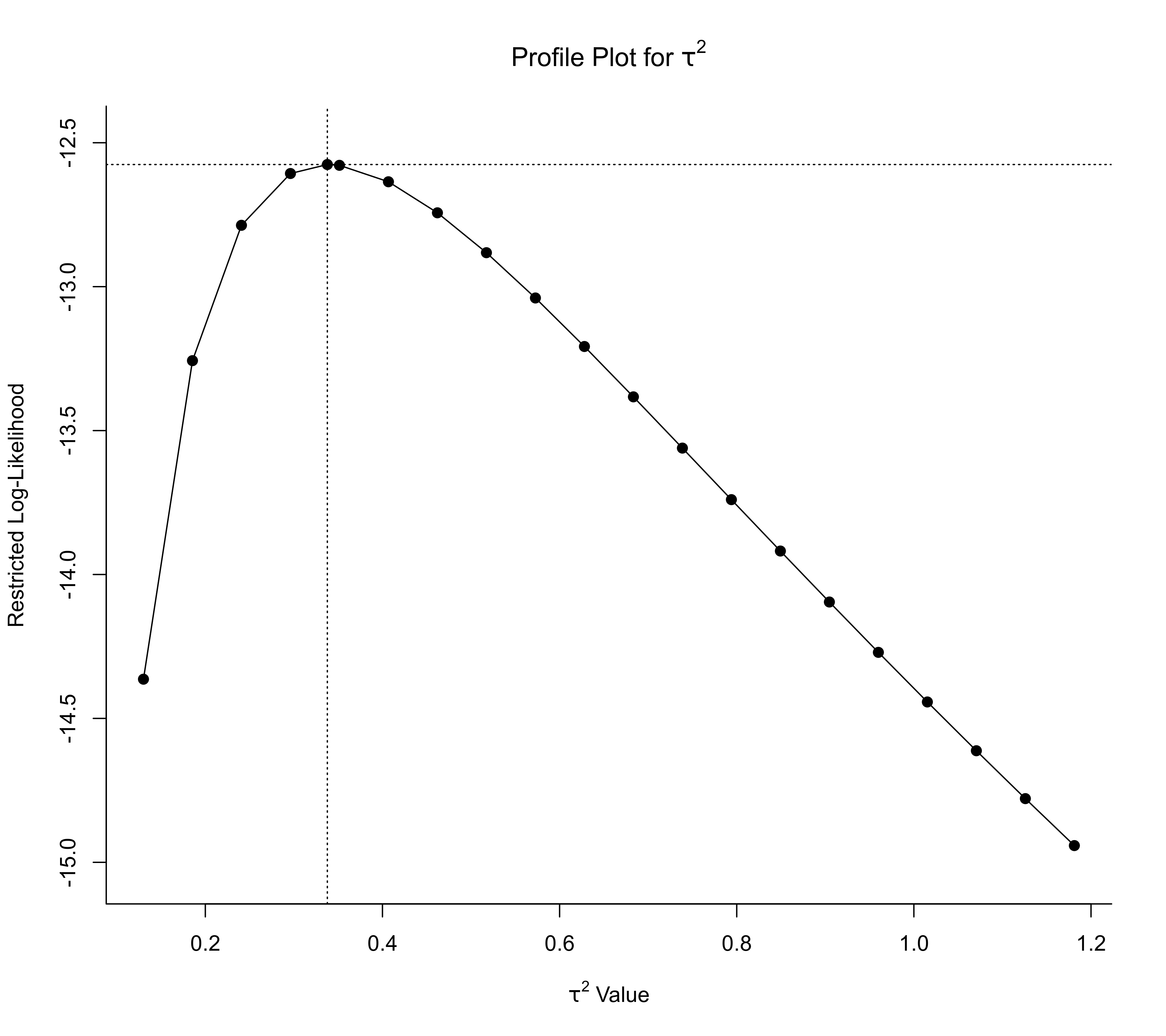 ### adjust xlim
profile(res, progbar=FALSE, xlim=c(0,1))
### adjust xlim
profile(res, progbar=FALSE, xlim=c(0,1))
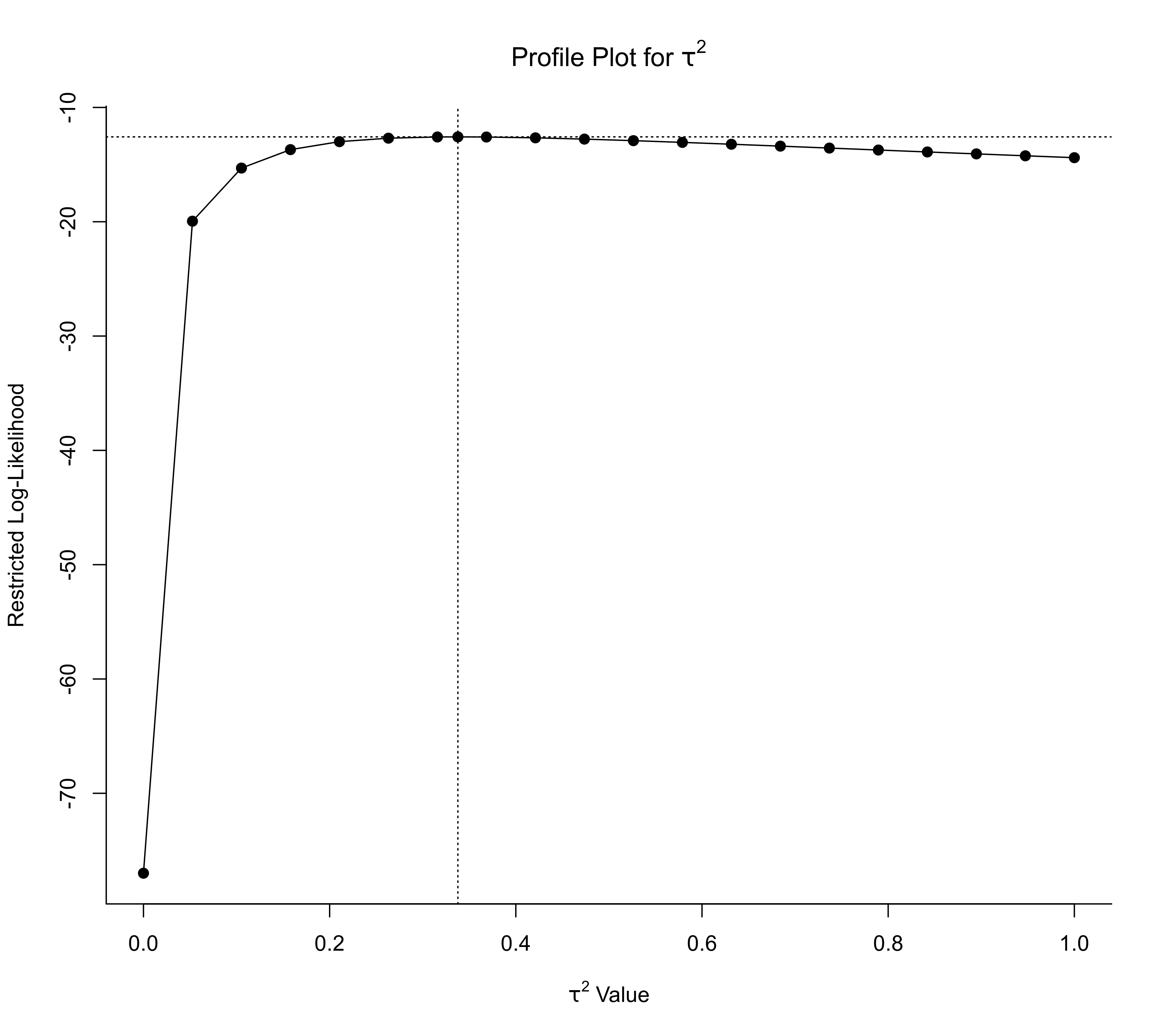 ### specify tau^2 values at which to profile the likelihood
profile(res, progbar=FALSE, steps=c(seq(0,0.2,length=20),seq(0.3,1,by=0.1)))
### specify tau^2 values at which to profile the likelihood
profile(res, progbar=FALSE, steps=c(seq(0,0.2,length=20),seq(0.3,1,by=0.1)))
 ### change data into long format
dat.long <- to.long(measure="OR", ai=tpos, bi=tneg, ci=cpos, di=cneg, data=dat.bcg, append=FALSE)
### set levels/labels for group ("con" = control/non-vaccinated, "exp" = experimental/vaccinated)
dat.long$group <- factor(dat.long$group, levels=c(2,1), labels=c("con","exp"))
### calculate log odds and corresponding sampling variances
dat.long <- escalc(measure="PLO", xi=out1, mi=out2, data=dat.long)
dat.long
#>
#> study group out1 out2 yi vi
#> 1 1 exp 4 119 -3.3928 0.2584
#> 2 1 con 11 128 -2.4541 0.0987
#> 3 2 exp 6 300 -3.9120 0.1700
#> 4 2 con 29 274 -2.2458 0.0381
#> 5 3 exp 3 228 -4.3307 0.3377
#> 6 3 con 11 209 -2.9444 0.0957
#> 7 4 exp 62 13536 -5.3860 0.0162
#> 8 4 con 248 12619 -3.9295 0.0041
#> 9 5 exp 33 5036 -5.0279 0.0305
#> 10 5 con 47 5761 -4.8087 0.0215
#> 11 6 exp 180 1361 -2.0230 0.0063
#> 12 6 con 372 1079 -1.0649 0.0036
#> 13 7 exp 8 2537 -5.7593 0.1254
#> 14 7 con 10 619 -4.1255 0.1016
#> 15 8 exp 505 87886 -5.1592 0.0020
#> 16 8 con 499 87892 -5.1713 0.0020
#> 17 9 exp 29 7470 -5.5514 0.0346
#> 18 9 con 45 7232 -5.0796 0.0224
#> 19 10 exp 17 1699 -4.6046 0.0594
#> 20 10 con 65 1600 -3.2034 0.0160
#> 21 11 exp 186 50448 -5.6030 0.0054
#> 22 11 con 141 27197 -5.2621 0.0071
#> 23 12 exp 5 2493 -6.2118 0.2004
#> 24 12 con 3 2338 -6.6584 0.3338
#> 25 13 exp 27 16886 -6.4384 0.0371
#> 26 13 con 29 17825 -6.4211 0.0345
#>
### fit bivariate random-effects model using rma.mv()
res <- rma.mv(yi, vi, mods = ~ group, random = ~ group | study, struct="UN", data=dat.long)
res
#>
#> Multivariate Meta-Analysis Model (k = 26; method: REML)
#>
#> Variance Components:
#>
#> outer factor: study (nlvls = 13)
#> inner factor: group (nlvls = 2)
#>
#> estim sqrt k.lvl fixed level
#> tau^2.1 2.6173 1.6178 13 no con
#> tau^2.2 1.5486 1.2444 13 no exp
#>
#> rho.con rho.exp con exp
#> con 1 - 13
#> exp 0.9450 1 no -
#>
#> Test for Residual Heterogeneity:
#> QE(df = 24) = 5270.3863, p-val < .0001
#>
#> Test of Moderators (coefficient 2):
#> QM(df = 1) = 15.5470, p-val < .0001
#>
#> Model Results:
#>
#> estimate se zval pval ci.lb ci.ub
#> intrcpt -4.0960 0.4529 -9.0432 <.0001 -4.9837 -3.2082 ***
#> groupexp -0.7414 0.1880 -3.9430 <.0001 -1.1099 -0.3729 ***
#>
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
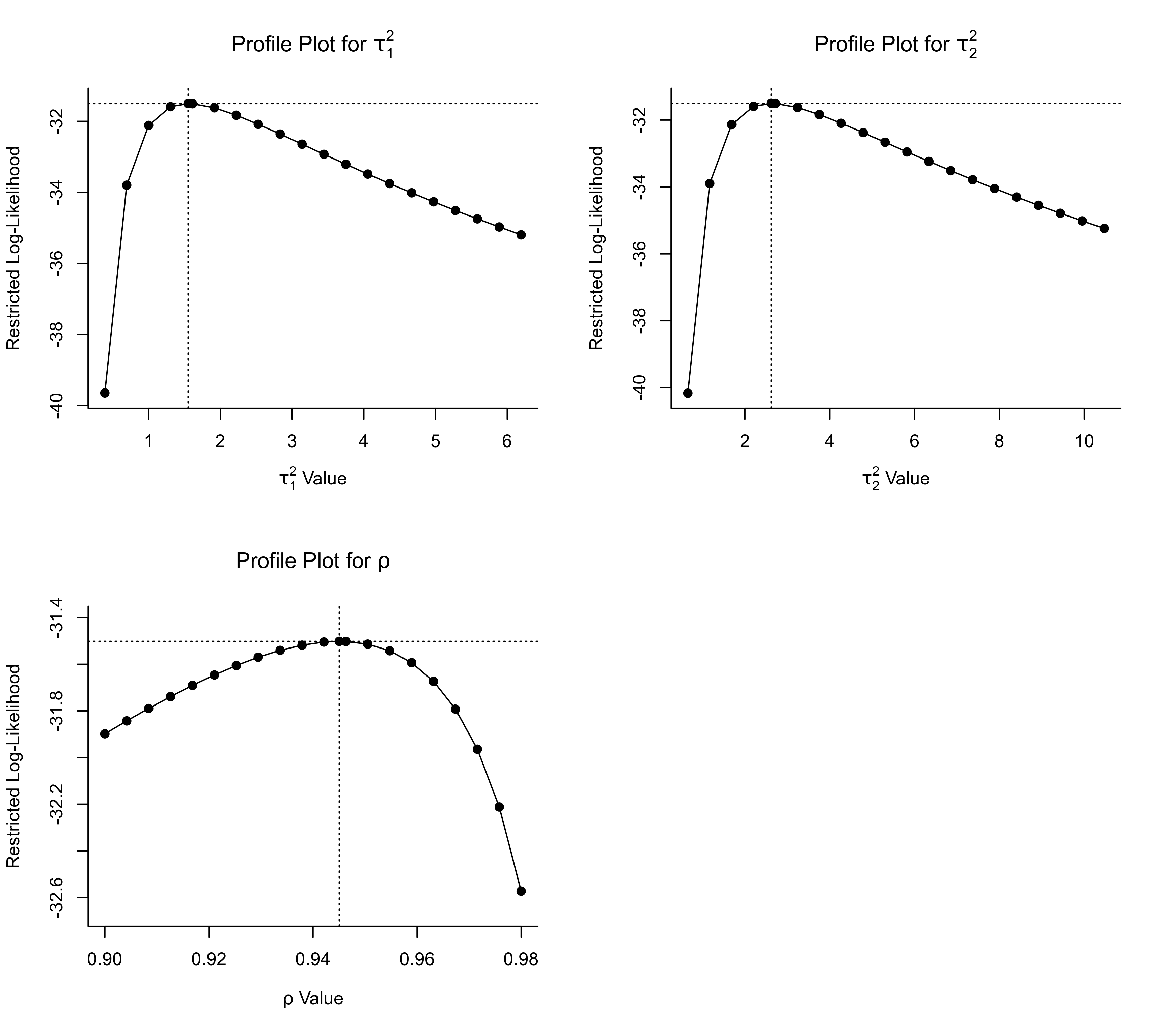
### profile over tau^2_1, tau^2_2, and rho
### note: for rho, adjust region over which profiling is done ('zoom in' on area around estimate)
par(mfrow=c(2,2))
profile(res, tau2=1)
profile(res, tau2=2)
profile(res, rho=1, xlim=c(0.90, 0.98))
par(mfrow=c(1,1))
### change data into long format
dat.long <- to.long(measure="OR", ai=tpos, bi=tneg, ci=cpos, di=cneg, data=dat.bcg, append=FALSE)
### set levels/labels for group ("con" = control/non-vaccinated, "exp" = experimental/vaccinated)
dat.long$group <- factor(dat.long$group, levels=c(2,1), labels=c("con","exp"))
### calculate log odds and corresponding sampling variances
dat.long <- escalc(measure="PLO", xi=out1, mi=out2, data=dat.long)
dat.long
#>
#> study group out1 out2 yi vi
#> 1 1 exp 4 119 -3.3928 0.2584
#> 2 1 con 11 128 -2.4541 0.0987
#> 3 2 exp 6 300 -3.9120 0.1700
#> 4 2 con 29 274 -2.2458 0.0381
#> 5 3 exp 3 228 -4.3307 0.3377
#> 6 3 con 11 209 -2.9444 0.0957
#> 7 4 exp 62 13536 -5.3860 0.0162
#> 8 4 con 248 12619 -3.9295 0.0041
#> 9 5 exp 33 5036 -5.0279 0.0305
#> 10 5 con 47 5761 -4.8087 0.0215
#> 11 6 exp 180 1361 -2.0230 0.0063
#> 12 6 con 372 1079 -1.0649 0.0036
#> 13 7 exp 8 2537 -5.7593 0.1254
#> 14 7 con 10 619 -4.1255 0.1016
#> 15 8 exp 505 87886 -5.1592 0.0020
#> 16 8 con 499 87892 -5.1713 0.0020
#> 17 9 exp 29 7470 -5.5514 0.0346
#> 18 9 con 45 7232 -5.0796 0.0224
#> 19 10 exp 17 1699 -4.6046 0.0594
#> 20 10 con 65 1600 -3.2034 0.0160
#> 21 11 exp 186 50448 -5.6030 0.0054
#> 22 11 con 141 27197 -5.2621 0.0071
#> 23 12 exp 5 2493 -6.2118 0.2004
#> 24 12 con 3 2338 -6.6584 0.3338
#> 25 13 exp 27 16886 -6.4384 0.0371
#> 26 13 con 29 17825 -6.4211 0.0345
#>
### fit bivariate random-effects model using rma.mv()
res <- rma.mv(yi, vi, mods = ~ group, random = ~ group | study, struct="UN", data=dat.long)
res
#>
#> Multivariate Meta-Analysis Model (k = 26; method: REML)
#>
#> Variance Components:
#>
#> outer factor: study (nlvls = 13)
#> inner factor: group (nlvls = 2)
#>
#> estim sqrt k.lvl fixed level
#> tau^2.1 2.6173 1.6178 13 no con
#> tau^2.2 1.5486 1.2444 13 no exp
#>
#> rho.con rho.exp con exp
#> con 1 - 13
#> exp 0.9450 1 no -
#>
#> Test for Residual Heterogeneity:
#> QE(df = 24) = 5270.3863, p-val < .0001
#>
#> Test of Moderators (coefficient 2):
#> QM(df = 1) = 15.5470, p-val < .0001
#>
#> Model Results:
#>
#> estimate se zval pval ci.lb ci.ub
#> intrcpt -4.0960 0.4529 -9.0432 <.0001 -4.9837 -3.2082 ***
#> groupexp -0.7414 0.1880 -3.9430 <.0001 -1.1099 -0.3729 ***
#>
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
### profile over tau^2_1, tau^2_2, and rho
### note: for rho, adjust region over which profiling is done ('zoom in' on area around estimate)
par(mfrow=c(2,2))
profile(res, tau2=1)
profile(res, tau2=2)
profile(res, rho=1, xlim=c(0.90, 0.98))
par(mfrow=c(1,1))
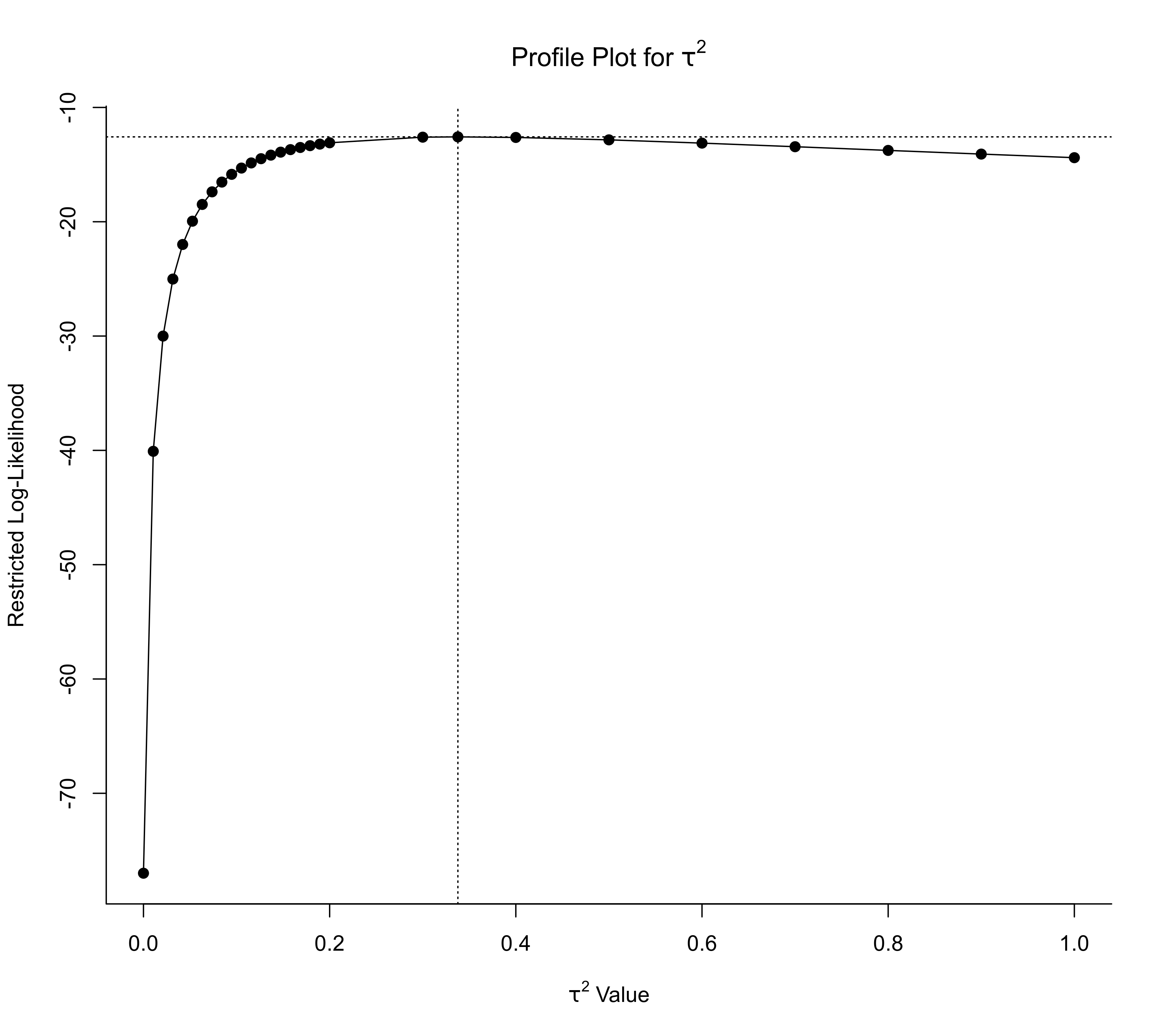
 ### an example where the peak of the likelihood profile is at 0
dat <- escalc(measure="RD", n1i=n1i, n2i=n2i, ai=ai, ci=ci, data=dat.hine1989)
res <- rma(yi, vi, data=dat)
profile(res, progbar=FALSE)
### an example where the peak of the likelihood profile is at 0
dat <- escalc(measure="RD", n1i=n1i, n2i=n2i, ai=ai, ci=ci, data=dat.hine1989)
res <- rma(yi, vi, data=dat)
profile(res, progbar=FALSE)