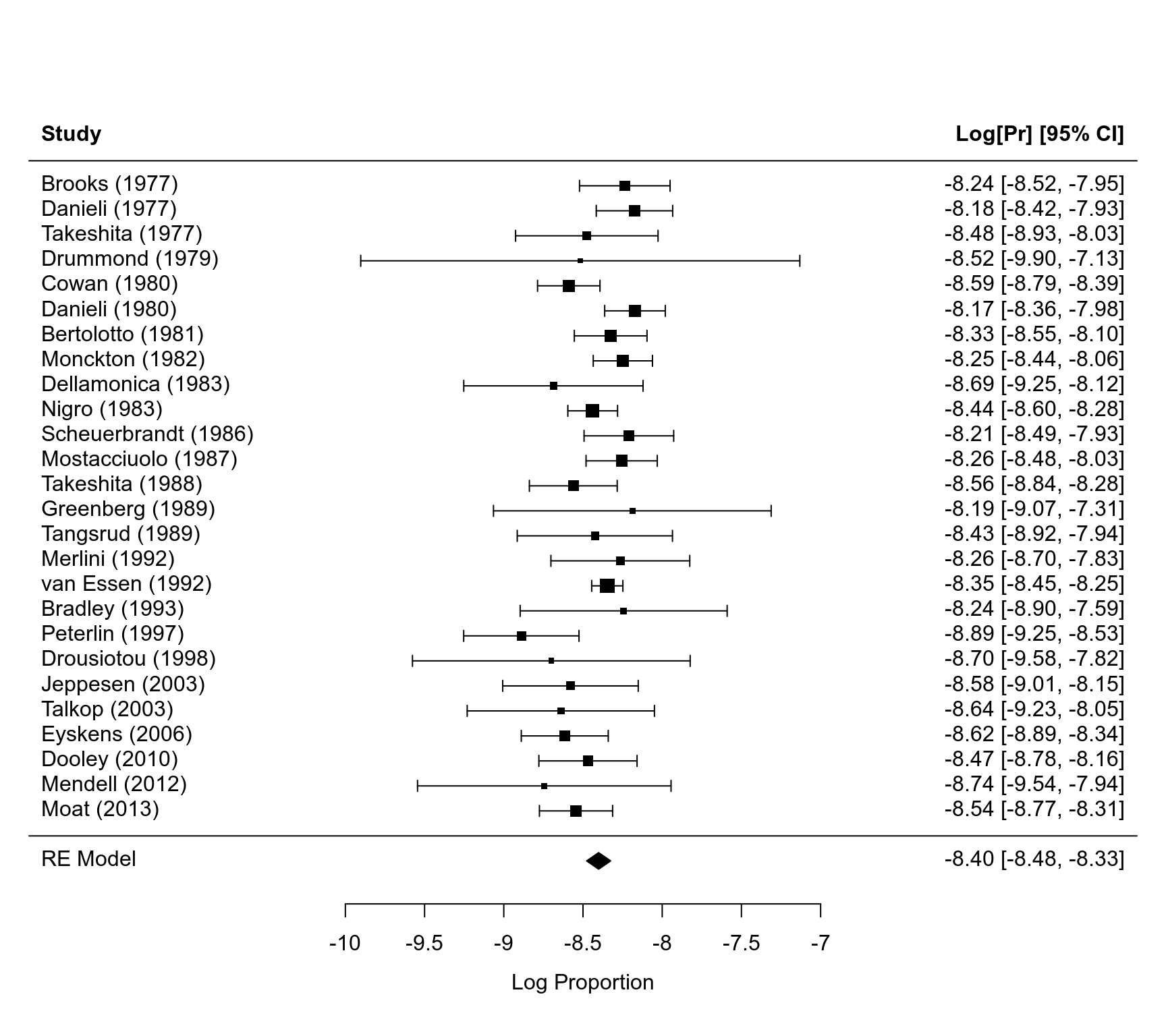
Duchenne Muscular Dystrophy (DMD) Prevalence Data
dat.crisafulli2020.Rd26 studies reporting estimates of the birth prevalence of Duchenne muscular dystrophy.
dat.crisafulli2020Format
The data frame contains the following columns:
| study | character | study label (first author, year) |
| pubyear | integer | publication year |
| country | factor | origin of investigated population |
| from, to | integer | time span of investigation (years) |
| cases | integer | number of DMD cases |
| total | integer | corresponding total population |
Details
Duchenne muscular dystrophy (DMD) is a rare disease that is caused by a genetic mutation and is characterized by impairment through muscle weakness and a reduced life expectancy.
Crisafulli et al. (2020) reported on a systematic review of data on the epidemiology of DMD, including estimates of the birth prevalence (which is of the order of a few per ten thousand). One of the originally reported studies (Koenig, 2019) is omitted here, as it constitutes an obvious outlier, and the reliability of the reported data is doubtful; Crisafulli et al. (2020) pointed out that “Concerning birth prevalence, Koenig et al. were found to be outliers. This study had problems with data collection in the last study year, as due to privacy issues, DMD cases were under-reported.”
Source
Crisafulli, S., Sultana, J., Fontana, A., Salvo, F., Messina, S., & Trifiro, G. (2020). Global epidemiology of Duchenne muscular dystrophy: an updated systematic review and meta-analysis. Orphanet Journal of Rare Diseases, 15, 141. https://doi.org/10.1186/s13023-020-01430-8
Concepts
medicine, epidemiology, proportions
Examples
# show (some) data
head(dat.crisafulli2020)
#> study pubyear country from to cases total
#> 1 Brooks (1977) 1977 UK 1953 1968 47 177413
#> 2 Danieli (1977) 1977 IT 1952 1972 66 234396
#> 3 Takeshita (1977) 1977 JP 1956 1970 19 91157
#> 4 Drummond (1979) 1979 NZ NA NA 2 10000
#> 5 Cowan (1980) 1980 AU 1960 1971 99 532302
#> 6 Danieli (1980) 1980 IT 1952 1972 105 371698
# compute logarithmic proportions and associated standard errors
library(metafor)
logp <- escalc(measure="PLN",
xi=cases, ni=total, slab=study,
data=dat.crisafulli2020)
# perform meta-analysis
rma01 <- rma.uni(logp)
# show results
rma01
#>
#> Random-Effects Model (k = 26; tau^2 estimator: REML)
#>
#> tau^2 (estimated amount of total heterogeneity): 0.0130 (SE = 0.0094)
#> tau (square root of estimated tau^2 value): 0.1139
#> I^2 (total heterogeneity / total variability): 41.79%
#> H^2 (total variability / sampling variability): 1.72
#>
#> Test for Heterogeneity:
#> Q(df = 25) = 37.4118, p-val = 0.0527
#>
#> Model Results:
#>
#> estimate se zval pval ci.lb ci.ub
#> -8.4014 0.0386 -217.6251 <.0001 -8.4770 -8.3257 ***
#>
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
# illustrate in a forest plot
forest(rma01, xlim=c(-12,-5))