Studies on Treatments for Chronic Obstructive Pulmonary Disease
dat.woods2010.RdResults from 3 trials examining the mortality risk of three treatments and placebo in patients with chronic obstructive pulmonary disease.
dat.woods2010Format
The data frame contains the following columns:
| author | character | first author / study name |
| treatment | character | treatment |
| r | integer | number of deaths |
| N | integer | number of patients |
Details
Count mortality statistics in randomised controlled trials of treatments for chronic obstructive pulmonary disease (Woods et al., 2010, Table 1).
Source
Woods, B. S., Hawkins, N., & Scott, D. A. (2010). Network meta-analysis on the log-hazard scale, combining count and hazard ratio statistics accounting for multi-arm trials: A tutorial. BMC Medical Research Methodology, 10, 54. https://doi.org/10.1186/1471-2288-10-54
Concepts
medicine, odds ratios, network meta-analysis
Examples
### Show full dataset
dat.woods2010
#> author treatment r N
#> 1 Boyd 1997 Salmeterol 1 229
#> 2 Boyd 1997 Placebo 1 227
#> 3 Calverly 2003 Fluticasone 4 374
#> 4 Calverly 2003 Salmeterol 3 372
#> 5 Calverly 2003 SFC 2 358
#> 6 Calverly 2003 Placebo 7 361
#> 7 Celli 2003 Salmeterol 1 554
#> 8 Celli 2003 Placebo 2 270
### Load netmeta package
suppressPackageStartupMessages(library(netmeta))
### Print odds ratios and confidence limits with two digits
oldset <- settings.meta(digits = 2)
### Change appearance of confidence intervals
cilayout("(", "-")
### Transform data from long arm-based format to contrast-based
### format. Argument 'sm' has to be used for odds ratio as summary
### measure; by default the risk ratio is used in the metabin function
### called internally.
pw <- pairwise(treatment, event = r, n = N,
studlab = author, data = dat.woods2010, sm = "OR")
pw
#> studlab treat1 treat2 TE seTE event1 n1 event2 n2 incr1 incr2
#> 1 Boyd 1997 Salmeterol Placebo -0.00881063 1.4173252 1 229 1 227 0 0
#> 2 Calverly 2003 Fluticasone Placebo -0.60382188 0.6311772 4 374 7 361 0 0
#> 3 Calverly 2003 Fluticasone SFC 0.65457491 0.8692018 4 374 2 358 0 0
#> 4 Calverly 2003 Fluticasone Salmeterol 0.28497571 0.7672979 4 374 3 372 0 0
#> 5 Calverly 2003 Salmeterol Placebo -0.88879759 0.6940644 3 372 7 361 0 0
#> 6 Calverly 2003 Salmeterol SFC 0.36959919 0.9158888 3 372 2 358 0 0
#> 7 Calverly 2003 SFC Placebo -1.25839679 0.8052894 2 358 7 361 0 0
#> 8 Celli 2003 Salmeterol Placebo -1.41751820 1.2270043 1 554 2 270 0 0
#> author treatment1 treatment2 r1 r2 N1 N2
#> 1 Boyd 1997 Salmeterol Placebo 1 1 229 227
#> 2 Calverly 2003 Fluticasone Placebo 4 7 374 361
#> 3 Calverly 2003 Fluticasone SFC 4 2 374 358
#> 4 Calverly 2003 Fluticasone Salmeterol 4 3 374 372
#> 5 Calverly 2003 Salmeterol Placebo 3 7 372 361
#> 6 Calverly 2003 Salmeterol SFC 3 2 372 358
#> 7 Calverly 2003 SFC Placebo 2 7 358 361
#> 8 Celli 2003 Salmeterol Placebo 1 2 554 270
### Conduct network meta-analysis
net <- netmeta(pw)
net
#> Number of studies: k = 3
#> Number of pairwise comparisons: m = 8
#> Number of observations: o = 2745
#> Number of treatments: n = 4
#> Number of designs: d = 2
#>
#> Common effects model
#>
#> Treatment estimate (other treatments vs 'Fluticasone'):
#> OR 95%-CI z p-value
#> Fluticasone . . . .
#> Placebo 1.81 (0.54-6.10) 0.96 0.3355
#> SFC 0.52 (0.09-2.85) -0.75 0.4514
#> Salmeterol 0.77 (0.19-3.08) -0.37 0.7078
#>
#> Random effects model
#>
#> Treatment estimate (other treatments vs 'Fluticasone'):
#> OR 95%-CI z p-value
#> Fluticasone . . . .
#> Placebo 1.81 (0.54-6.10) 0.96 0.3355
#> SFC 0.52 (0.09-2.85) -0.75 0.4514
#> Salmeterol 0.77 (0.19-3.08) -0.37 0.7078
#>
#> Quantifying heterogeneity / inconsistency:
#> tau^2 = 0; tau = 0; I^2 = 0% (0.0%-89.6%)
#>
#> Tests of heterogeneity (within designs) and inconsistency (between designs):
#> Q d.f. p-value
#> Total 0.57 2 0.7525
#> Within designs 0.56 1 0.4524
#> Between designs 0.00 1 0.9485
#>
#> Details of network meta-analysis methods:
#> - Frequentist graph-theoretical approach
#> - DerSimonian-Laird estimator for tau^2
#> - Calculation of I^2 based on Q
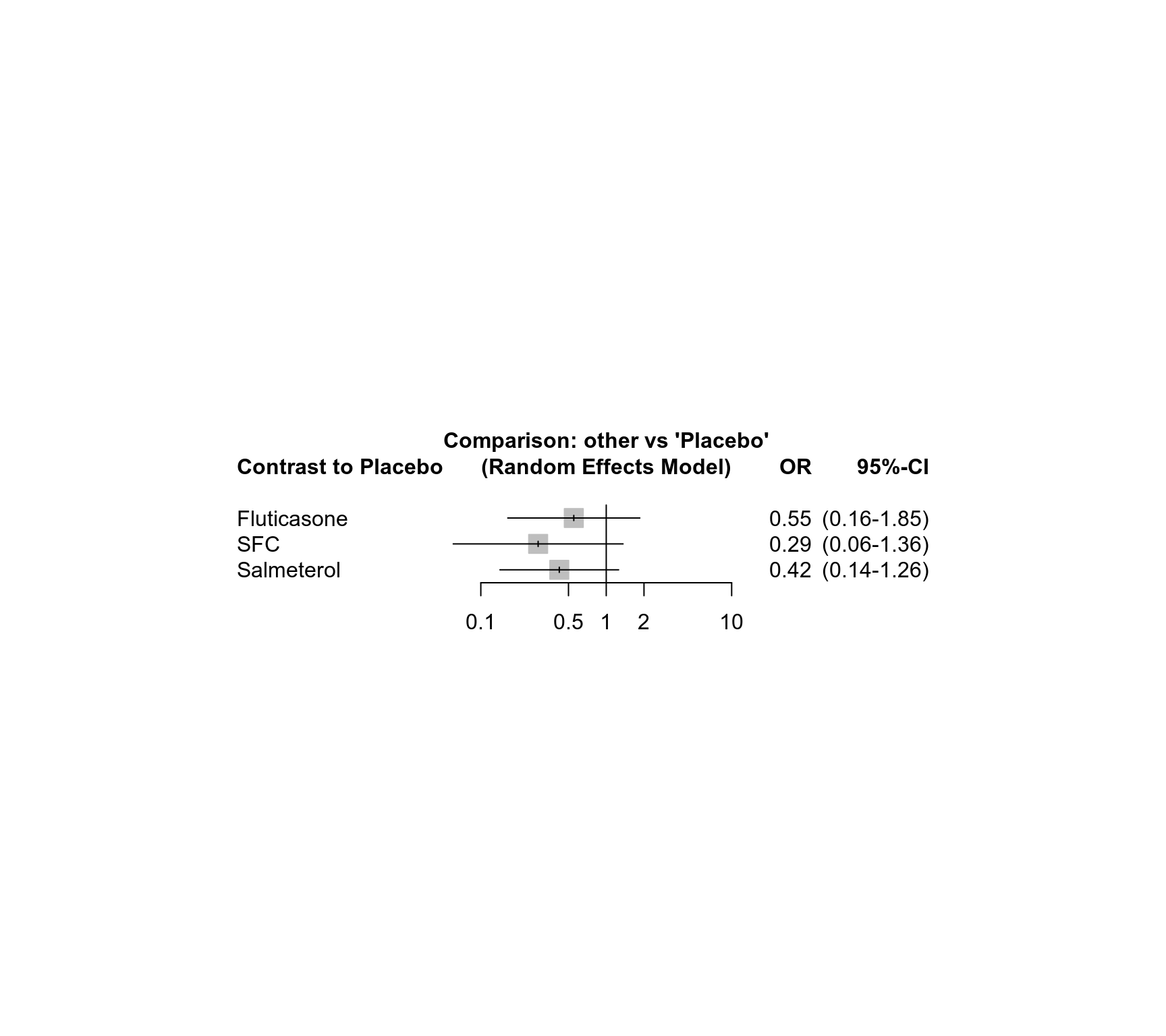
### Show forest plot
forest(net, ref = "Placebo", drop = TRUE,
leftlabs = "Contrast to Placebo")
 ### Use previous settings
settings.meta(oldset)
### Use previous settings
settings.meta(oldset)